Source/Disclosures
Published by:
Hydes T, et al. Abstract OS048. Presented at: International Liver Congress; June 22-26, 2022; London (hybrid meeting).
Disclosures:
Hydes reports no relevant financial disclosures.
LONDON — Elevated noninvasive markers of liver fibrosis correlated with an increased risk for cardiovascular events, end-stage renal disease and worse survival in patients with chronic kidney disease and nonalcoholic fatty liver disease.
“Multimorbidity is increasing, and it is vital to understand the clinical consequences of having more than one medical condition,” Theresa Hydes, MBBS, BSc, PhD, NIHR clinical lecturer in hepatology at the University of Liverpool, told Healio. “Fatty liver disease, in particular, is independently associated with several non-liver conditions, including heart disease and chronic kidney disease.”

Seeking to assess the effect of NAFLD and NAFLD fibrosis on adverse clinical outcomes and mortality among patients with chronic kidney disease (CKD), Hydes and colleagues analyzed data from 26,074 patients using the UK Biobank. Participants provided information related to medical history, demographics and lifestyle factors, which was supplemented via electronic linkage to hospital records and death records.
Researchers used Cox regression to estimate hazard ratios associated with NAFLD and advanced liver fibrosis on CV events, progression to end-stage renal disease (ESRD) and all-cause mortality.
At baseline, 54,5% of patients with CKD had NAFLD, with evidence of advanced fibrosis among 7% [NAFLD fibrosis score (NFS) 0.676], 3.2% [elevated fibrosis-4 (FIB-4) > 2.67] and 1.1% [AST to platelet ratio index (APRI) 1].
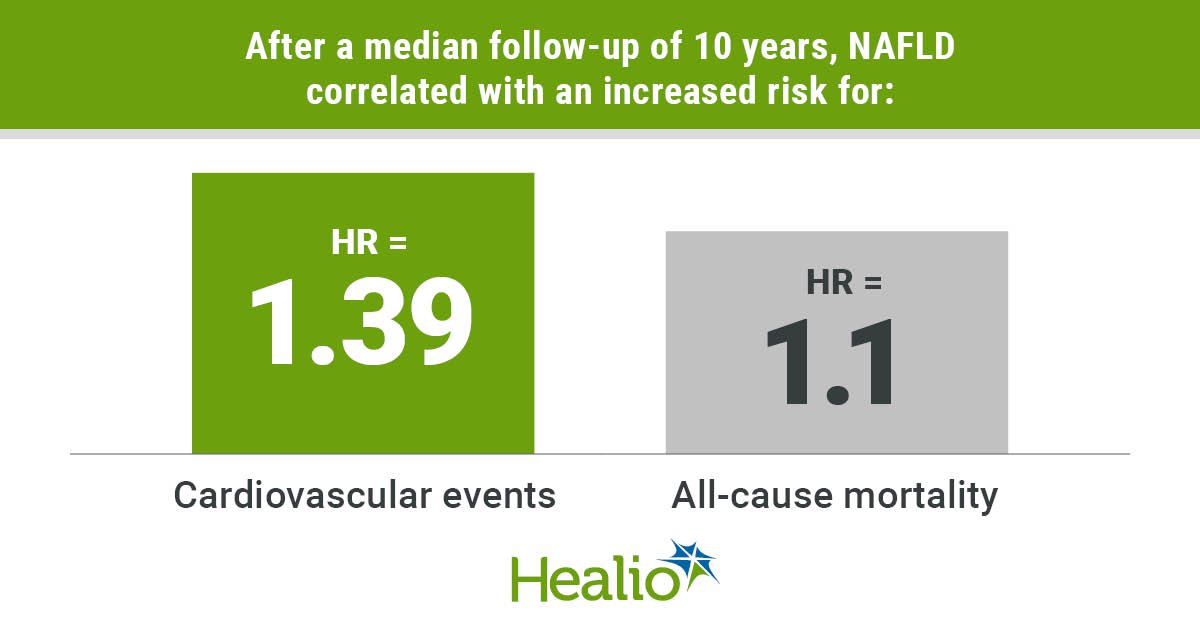
After a median follow-up of 10 years, NAFLD correlated with an increased risk for CV events (HR = 1.39; 95% CI, 1.29-1.51) and all-cause mortality (HR = 1.1; 95% CI, 1.01-1.19) but not ESRD (HR = 1.22; 95% CI, 0.95-1.56) in a univariate analysis. Following multivariate adjustment for demographics, metabolic factors and baseline renal functions, NAFLD did not associate with an increased risk for primary outcomes.
Advanced liver fibrosis using all scores correlated with an increased risk for all-cause mortality (HR = 2.34-2.9), and NFS and FIB-4 associated with an elevated risk for CV events (HR = 2.49; 95% CI, 2.11-2.93 and HR = 1.94; 95% CI, 1.53-2.45) and ESRD (HR = 6.85; 95% CI, 4.29-10.94 and HR = 2.35; 95% CI, 1.19-4.67). After full adjustment, FIB-4 correlated with an increased incidence of CV events (HR = 1.39; 95% CI, 1.06-1.82), notably heart failure (HR = 1.65; 95% CI, 1.16-2.33).
Both FIB-4 and APRI associated with all-cause mortality (HR = 1.55; 95% CI, 1.21-2 and HR = 2.83; 95% CI, 1.95-4.11) and NFS ( –1.455) associated with progression to ESRD (HR = 1.89; 95% CI, 1.13-3.17).
“These results highlight the importance of enhanced recognition of fatty liver disease with fibrosis in people with chronic kidney disease to inform the need for vigorous cardiometabolic risk factor control in this group,” Hydes said. “It also suggests the need for work to understand the mechanisms linking these conditions to help drive new drug discoveries.”






